Load data and libraries
library(tidyverse)
library(patchwork)
library(paletteer)
tuesdata <- tidytuesdayR::tt_load(2025, week = 35)
frogID_data <- tuesdata$frogID_data
frog_names <- tuesdata$frog_namesSeptember 2, 2025
This week’s data is from the FrogID dataset, which I sourced and curated! The dataset contains 6 years worth of frog recordings. To meet file size requirements, I limited the Tidy Tuesday dataset to just 2023 data.
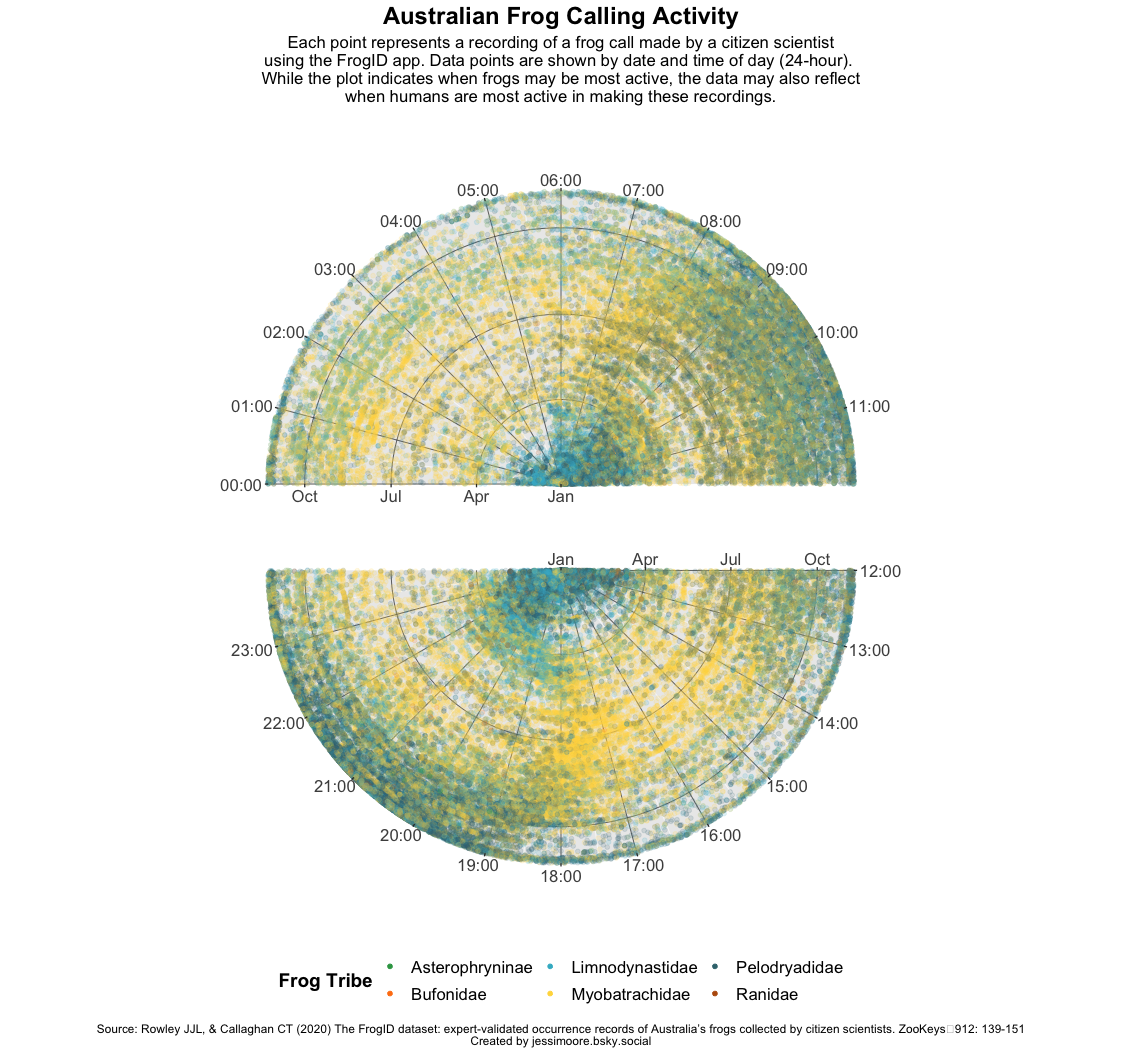
I made a relatively quick plot this week to try to bump up my efficiency. Not a lot of info can be gleaned from the tribe colours, apart from that Pelodryadidae (Treefrogs) appear to be the most common, followed by Myobatrachidae (Ground/water frogs). The seasonal and time-of-day patterns are interesting and to be expected, although I wonder how much these are influenced by when people are most likely to be outside and recording frogs.
frog_names[frog_names == '—'] <- NA
frogs <- frogID_data %>%
left_join(frog_names, by = "scientificName") %>%
select(c(scientificName, eventDate, eventTime, commonName, stateProvince,
decimalLatitude, decimalLongitude, tribe)) %>%
drop_na(commonName)
frogs_am <- frogs %>%
filter(eventTime < hms("12:00:00"))
frogs_pm <- frogs %>%
filter(eventTime >= hms("12:00:00"))am <- ggplot(frogs_am, aes(eventTime, eventDate, color = tribe)) +
geom_point(alpha = 0.15) +
scale_x_time(breaks = seq(0, 23 * 3600, by = 3600),
labels = function(x) strftime(x, "%H:%M")) +
scale_color_paletteer_d("ggthemes::Classic_Green_Orange_6") +
coord_radial(expand = FALSE, r.axis.inside = TRUE,
start = -0.5*pi, end = 0.5*pi) +
guides(color = guide_legend(override.aes = list(alpha = 1))) +
labs(x = NULL, y = NULL, color = "Frog Tribe")
pm <- ggplot(frogs_pm, aes(eventTime, eventDate, color = tribe)) +
geom_point(alpha = 0.15) +
scale_x_time(breaks = seq(0, 23 * 3600, by = 3600),
labels = function(x) strftime(x, "%H:%M")) +
scale_color_paletteer_d("ggthemes::Classic_Green_Orange_6") +
coord_radial(expand = FALSE, r.axis.inside = TRUE,
start = 0.5*pi, end = -0.5*pi) +
guides(color = guide_legend(override.aes = list(alpha = 1))) +
labs(x = NULL, y = NULL, color = "Frog Tribe")
combined_plot <- am / pm + plot_layout(guides = "collect") +
plot_annotation(
title = "Australian Frog Calling Activity",
subtitle = glue::glue("Each point represents a recording of a frog call made by a citizen scientist
using the FrogID app. Data points are shown by date and time of day (24-hour).
While the plot indicates when frogs may be most active, the data may also reflect
when humans are most active in making these recordings."),
caption = glue::glue("Source: Rowley JJL, & Callaghan CT (2020) The FrogID dataset: expert-validated occurrence records of Australia’s frogs collected by citizen scientists. ZooKeys 912: 139-151
Created by jessimoore.bsky.social")
) &
theme(axis.text = element_text(size = 14),
panel.grid.major = element_line(color = "grey25", linewidth = 0.3),
legend.key = element_blank(),
plot.title = element_text(size = 20, face = "bold", hjust = 0.5),
plot.subtitle = element_text(size = 14, hjust = 0.5),
legend.title = element_text(size = 16, face = "bold"),
legend.text = element_text(size = 14),
legend.position = "bottom",
plot.caption = element_text(size = 10, hjust = 0.5),
plot.caption.position = "plot")